Transcription is the key step during differential gene expression, which underlies all biological processes including animal development. Transcription is tightly regulated in a highly dynamic fashion via two DNA sequence elements, enhancers and core-promoters. Enhancers encode the spatiotemporal information for transcription, containing short DNA sequence motifs to which transcription factors bind and subsequently recruit cofactors. Core-promoters are ~100bp short sequences surrounding the transcription start sites (TSSs), at which the RNA polymerase II machinery assembles. A key function of core-promoters is to respond rapidly and strongly towards enhancers, efficiently converting enhancer activity into productive transcription.
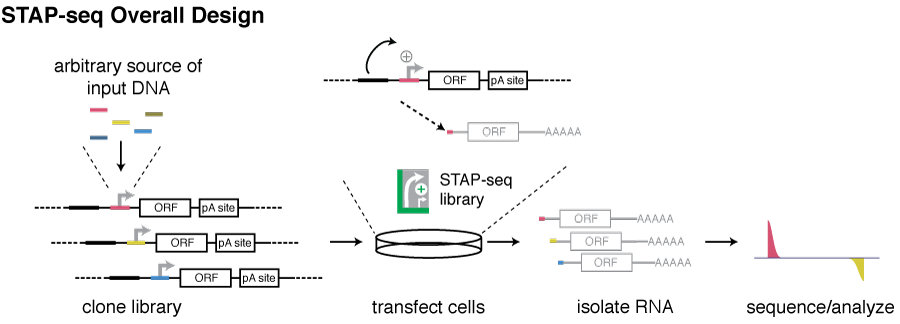
As this enhancer responsiveness cannot be assessed from endogenous transcription rates and needs to be measured in standardized assays, we have developed Self-Transcribing Active core-Promoter sequencing (STAP-seq). STAP-seq is a high throughput technique to quantify the sequence-intrinsic capacity to initiate transcription provided an enhancer for millions of candidate DNA fragments from arbitrary sources.
Candidate DNA fragments are cloned at the position of the core promoter (also called minimal promoter) of a transcriptional reporter, namely downstream of an enhancer and upstream of a reporter gene. Transcription initiation will occur within candidates that can function as core promoters, leading to the production of chimeric reporter transcripts that start with short 5’ tags derived from corresponding the candidate fragments and are otherwise identical. Deep sequencing of these tags enables the quantification of the initiation capacity, as well as the identification of the initiation positions.
What are the features of STAP-seq?
STAP-seq is ectopic and plasmid-based and transcription initiation is assessed in a constant environment with a single enhancer and without any influence of the candidates’ endogenous genomic loci or chromatin states. The measured activity and the identified transcription initiation site (TSS) therefore directly reflect the sequence-intrinsic transcription capacity of the candidate sequences in an orientation-specific manner. Tests with different enhancers and in two different Drosophila cell lines indeed show that the enhancer responsiveness measured by STAP-seq is independent of the cell type used and consistent across different developmental enhancers.
—
