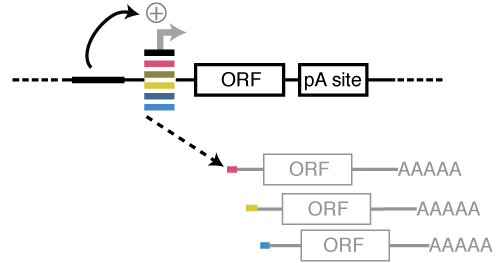
Self-Transcribing Active core-Promoter sequencing (STAP-seq), is a high throughput method that quantifies the sequence-intrinsic capacity to initiate transcription provided a single defined enhancer for millions of candidate DNA fragments from arbitrary sources. STAP-seq allows the determination of each candidate’s enhancer-responsiveness, i.e. the extent to which the enhancer input is converted into transcription activation, a functional property of core-promoters that is of central importance for differential gene expression. STAP-seq is complementary to STARR-seq: while STARR-seq measures enhancer activity using a single defined core-promoter, STAP-seq measures core-promoter activity using a single defined enhancer.
The reporter constructs contain the candidate fragments at the canonical position of the core-promoter (also called minimal promoter) between the enhancer and a reporter gene. Transcription initiation within candidate fragments leads to reporter transcripts that start with short 5’ sequence tags derived from the corresponding core-promoter candidate. Deep sequencing of these tags enables the quantification of the transcriptional activity and the identification of the transcription initiation sites (TSSs) for each candidate.